KofamScan-KEGG官方推荐的使用系同源和隐马尔可夫模型进行KO注释
文章目录
简介
KofamScan 是一款基于 KEGG 直系同源和隐马尔可夫模型(HMM)的基因功能注释工具,可通过同源搜索和预先计算的自适应分数阈值,将 KEGG 同源物(KOs)分配给蛋白质序列的隐马尔可夫模型(KOfam)数据库。在线版本可在 https://www.genome.jp/tools/kofamkoala/ 上获取。KofamKOALA 比现有的 KO 分配工具更快,其准确性可与性能最好的工具相媲美。KofamKOALA 的功能注释有助于将基因与 KEGG 通路图等 KEGG 资源联系起来,并促进分子网络的重建。
安装
# 软件
(base) [yut@io02 ~]$ mamba install -c bioconda kofamscan
# 数据库
# filezila打开ftp.genome.jp看一下最新版数据库更新时间
mkdir -p KoFamDB20231025
cd KoFamDB20231025
#数据库:
wget ftp://ftp.genome.jp/pub/db/kofam/ko_list.gz
wget ftp://ftp.genome.jp/pub/db/kofam/profiles.tar.gz
gunzip ko_list.gz
tar xvzf profiles.tar.gz
使用
输入蛋白序列
查询文件是包含一个或多个氨基酸序列的 FASTA 文件。不能使用核苷酸序列。每个序列必须有一个唯一的名称。序列名称是介于标题符号(“>”)和第一个空白字符(空格、制表符、换行符等)之间的字符串。请勿在">"之后使用空格。
(base) [yut@node01 TestData]$ grep -c ">" GCA_002343985.1_ASM234398v1_genomic.faa
2486 # 蛋白数量
输出detail-tsv格式
time exec_annotation -p ~/Database/KoFamDB20231025/profiles -k ~/Database/KoFamDB20231025/ko_list --cpu 10 --tmp-dir . -E 1e-5 -f detail-tsv -o GCA_002343985.1_ASM234398v1_genomic.faa_kofamscan_detail-tsv.tsv GCA_002343985.1_ASM234398v1_genomic.faa &>GCA_002343985.1_ASM234398v1_genomic.faa_kofamscan_detail.log
# -p与-k为上述数据库配置文件
# -E 为e-value
# -f为输出格式,包括:
-f, --format <format> Format of the output [detail]
detail: Detail for each hits (including hits below threshold)
detail-tsv: Tab separeted values for detail format
mapper: KEGG Mapper compatible format
mapper-one-line: Similar to mapper, but all hit KOs are listed in one line
# 3m18.871s
输出detail格式
(base) [yut@node01 TestData]$ time exec_annotation -p ~/Database/KoFamDB20231025/profiles -k ~/Database/KoFamDB20231025/ko_list --cpu 10 --tmp-dir . -E 1e-5 -f detail -o GCA_002343985.1_ASM234398v1_genomic.faa_kofamscan_detail_format.tsv GCA_002343985.1_ASM234398v1_genomic.faa &>GCA_002343985.1_ASM234398v1_genomic.faa_kofamscan_detail.log&
# real 3m0.691s
输出mapper格式
(base) [yut@node01 TestData]$ time exec_annotation -p ~/Database/KoFamDB20231025/profiles -k ~/Database/KoFamDB20231025/ko_list --cpu 10 --tmp-dir . -E 1e-5 -f mapper -o GCA_002343985.1_ASM234398v1_genomic.faa_kofamscan_mapper_format.tsv GCA_002343985.1_ASM234398v1_genomic.faa &>GCA_002343985.1_ASM234398v1_genomic.faa_kofamscan_mapper.log&
# real 3m13.254s
输出结果
detail和detail-tsv格式
(base) [yut@node01 TestData]$ head GCA_002343985.1_ASM234398v1_genomic.faa_kofamscan_detail-tsv.tsv
# gene name KO thrshld score E-value "KO definition"
# --------- ------ ------- ------ --------- -------------
* GLGNCOCG_00001 K09946 28.63 105.8 3.2e-31 "uncharacterized protein"
* GLGNCOCG_00002 K22441 90.53 175.7 2.4e-52 "diamine N-acetyltransferase [EC:2.3.1.57]"
GLGNCOCG_00002 K03789 105.10 66.4 4.4e-19 "[ribosomal protein S18]-alanine N-acetyltransferase [EC:2.3.1.266]"
GLGNCOCG_00002 K03828 117.57 66.1 4.1e-19 "putative acetyltransferase [EC:2.3.1.-]"
GLGNCOCG_00002 K03825 113.37 57.6 1.7e-16 "L-phenylalanine/L-methionine N-acetyltransferase [EC:2.3.1.53 2.3.1.-]"
GLGNCOCG_00002 K24217 175.13 55.7 6e-16 "L-amino acid N-acyltransferase [EC:2.3.1.-]"
GLGNCOCG_00002 K09994 117.73 50.8 2.6e-14 "(aminoalkyl)phosphonate N-acetyltransferase [EC:2.3.1.280]"
GLGNCOCG_00002 K15520 93.30 50.6 3.5e-14 "mycothiol synthase [EC:2.3.1.189]"
(base) [yut@node01 TestData]$ head GCA_002343985.1_ASM234398v1_genomic.faa_kofamscan_detail_format.tsv
# gene name KO thrshld score E-value KO definition
#-------------------- ------ ------- ------ --------- ---------------------
* GLGNCOCG_00001 K09946 28.63 105.8 3.2e-31 uncharacterized protein
* GLGNCOCG_00002 K22441 90.53 175.7 2.4e-52 diamine N-acetyltransferase [EC:2.3.1.57]
GLGNCOCG_00002 K03789 105.10 66.4 4.4e-19 [ribosomal protein S18]-alanine N-acetyltransferase [EC:2.3.1.266]
GLGNCOCG_00002 K03828 117.57 66.1 4.1e-19 putative acetyltransferase [EC:2.3.1.-]
GLGNCOCG_00002 K03825 113.37 57.6 1.7e-16 L-phenylalanine/L-methionine N-acetyltransferase [EC:2.3.1.53 2.3.1.-]
GLGNCOCG_00002 K24217 175.13 55.7 6e-16 L-amino acid N-acyltransferase [EC:2.3.1.-]
GLGNCOCG_00002 K09994 117.73 50.8 2.6e-14 (aminoalkyl)phosphonate N-acetyltransferase [EC:2.3.1.280]
GLGNCOCG_00002 K15520 93.30 50.6 3.5e-14 mycothiol synthase [EC:2.3.1.189]
(base) [yut@node01 TestData]$ sort -t $'\t' -k4 -n GCA_002343985.1_ASM234398v1_genomic.faa_kofamscan_detail-tsv.tsv|head
# --------- ------ ------- ------ --------- -------------
# gene name KO thrshld score E-value "KO definition"
GLGNCOCG_01738 K19152 0.5 1.9e+02 "small toxic protein IbsD"
GLGNCOCG_02049 K21664 1.7 4.3 "KSHV latency-associated nuclear antigen"
GLGNCOCG_02331 K21664 2.9 1.8 "KSHV latency-associated nuclear antigen"
GLGNCOCG_00788 K21664 4.7 0.53 "KSHV latency-associated nuclear antigen"
GLGNCOCG_01757 K19313 5.6 1.5 "aminocyclitol acetyltransferase [EC:2.3.-.-]"
GLGNCOCG_01062 K16402 6.8 0.023 "soraphen polyketide synthase B"
GLGNCOCG_00067 K16007 7.1 0.033 "8,8a-deoxyoleandolide synthase 1"
GLGNCOCG_01945 K26748 7.2 0.27 "MFS transporter, AAHS family, 2,4-dichlorophenoxyacetate transporter"
# 有明确KO号的基因数量
(base) [yut@node01 TestData]$ awk '$1~/*/' GCA_002343985.1_ASM234398v1_genomic.faa_kofamscan_detail-tsv.tsv|wc -l
1216
(base) [yut@node01 TestData]$ echo|awk '{print 1216/2486*100}'
48.9139
可以看到detailj及detail-tsv格式是用制表符将所有比对的结果都输出,即除了满足阈值< 1e-5,同时也将大于该阈值的结果输出。总共7列,第一列是标记该基因是否分配到Knumber,带星号的表示有Knumber;第二列表示输入基因名称;第三列是KO号;第四列是阈值;第五列是得分,得分一般大于前面阈值列的就会有KO;第六列是E-value;第七列是KO definition。该基因组最终有48.91%的蛋白序列有KO注释。
mapper格式
(base) [yut@node01 TestData]$ head GCA_002343985.1_ASM234398v1_genomic.faa_kofamscan_mapper_format.tsv
GLGNCOCG_00001 K09946
GLGNCOCG_00002 K22441
GLGNCOCG_00003
GLGNCOCG_00004 K03286
GLGNCOCG_00005
GLGNCOCG_00006
GLGNCOCG_00007
GLGNCOCG_00008
GLGNCOCG_00009
GLGNCOCG_00010
# 一个基因注释到多个KO
(base) [yut@node01 TestData]$ grep GLGNCOCG_00297 GCA_002343985.1_ASM234398v1_genomic.faa_kofamscan_mapper_format.tsv
GLGNCOCG_00297 K13990
GLGNCOCG_00297 K00603
GLGNCOCG_00297 K01500
(base) [yut@node01 TestData]$ grep GLGNCOCG_00297 GCA_002343985.1_ASM234398v1_genomic.faa_kofamscan_detail_format.tsv
* GLGNCOCG_00297 K13990 507.67 749.9 4.5e-226 glutamate formiminotransferase / formiminotetrahydrofolate cyclodeaminase [EC:2.1.2.5 4.3.1.4]
* GLGNCOCG_00297 K00603 405.93 411.8 4.9e-124 glutamate formiminotransferase / 5-formyltetrahydrofolate cyclo-ligase [EC:2.1.2.5 6.3.3.2]
* GLGNCOCG_00297 K01500 201.63 215.6 7.4e-65 methenyltetrahydrofolate cyclohydrolase [EC:3.5.4.9]
GLGNCOCG_00297 K01746 429.53 65.3 8.3e-19 formiminotetrahydrofolate cyclodeaminase [EC:4.3.1.4]
GLGNCOCG_00297 K10740 25.77 10.7 0.042 replication factor A3
# 有KO的
(base) [yut@node01 TestData]$ awk -F"\t" '$2!=""' GCA_002343985.1_ASM234398v1_genomic.faa_kofamscan_mapper_format.tsv|wc -l
1216
mapper格式总共两列,第一列是基因ID,第二列是Knumber,没有注释到的为空,注意一个基因可能同时注释到多个KO。
- mapper-one-line格式:类似上面的mapper格式,不一样的是一个基因如果有多个KO则以制表符分割放在同一行。
(base) [yut@node01 TestData]$ less GCA_002343985.1_ASM234398v1_genomic.faa_kofamscan_mapper-one-line_format.tsv
GLGNCOCG_00294 K07304
GLGNCOCG_00295 K01924
GLGNCOCG_00296 K01468
GLGNCOCG_00297 K13990 K00603 K01500
GLGNCOCG_00298
GLGNCOCG_00299
GLGNCOCG_00300
GLGNCOCG_00301
GLGNCOCG_00302
(base) [yut@node01 TestData]$ awk 'NF>2' GCA_002343985.1_ASM234398v1_genomic.faa_kofamscan_mapper-one-line_format.tsv|cat -A
GLGNCOCG_00069^IK13693^IK01079$
GLGNCOCG_00108^IK01417^IK20276$
GLGNCOCG_00297^IK13990^IK00603^IK01500$
GLGNCOCG_00486^IK03668^IK06079$
GLGNCOCG_00489^IK06079^IK03668$
GLGNCOCG_00776^IK00027^IK00029$
GLGNCOCG_00998^IK25153^IK25151$
常用命令
参数:evalue:1e-5(the ORFs were searched against the Kyoto Encyclopedia of Genes and Genomes (KEGG) database [48] using KofamScan version 1.2.0 (E-value < 10−5, score>predefined thresholds by KofamScan) [49]. 参考https://www.nature.com/articles/s41396-023-01546-2,https://microbiomejournal.biomedcentral.com/articles/10.1186/s40168-023-01621-y)
每行一个基因,一个基因多个KO则多行输出,不输出注释不到的基因,
(base) [yut@node01 TestData]$ time exec_annotation -p ~/Database/KoFamDB20231025/profiles -k ~/Database/KoFamDB20231025/ko_list --cpu 10 -E 1e-5 -f mapper --no-report-unannotated -o GCA_002343985.1_ASM234398v1_genomic.faa_kofamscan_mapper_annot_format_1e- 5.tsv GCA_002343985.1_ASM234398v1_genomic.faa &
tmp目录
- 不写 --tmp-dir则默认在当前目录创建tmp目录,其下生成tabular目录,里面是所有KO的HMM信息。
- 注意:如果使用
parallel同时并行多个文件时,一定要为每个指定一个不同名称的目录,否则则会互相影响,输出不完整。
(base) [yut@node01 TestData]$ ls tmp
tabular
(base) [yut@node01 TestData]$ ll tmp/tabular/|head
总用量 51M
-rw-r--r-- 1 yut lzdx 2.4K 11月 16 11:17 K00001
-rw-r--r-- 1 yut lzdx 1.2K 11月 16 11:17 K00002
-rw-r--r-- 1 yut lzdx 2.3K 11月 16 11:17 K00003
-rw-r--r-- 1 yut lzdx 2.9K 11月 16 11:17 K00004
-rw-r--r-- 1 yut lzdx 1.4K 11月 16 11:17 K00005
-rw-r--r-- 1 yut lzdx 1.2K 11月 16 11:17 K00006
-rw-r--r-- 1 yut lzdx 997 11月 16 11:17 K00007
-rw-r--r-- 1 yut lzdx 2.8K 11月 16 11:17 K00008
-rw-r--r-- 1 yut lzdx 1.2K 11月 16 11:17 K00009
(base) [yut@node01 TestData]$ head tmp/tabular/K00001
# --- full sequence ---- --- best 1 domain ---- --- domain number estimation ----
# target name accession query name accession E-value score bias E-value score bias exp reg clu ov env dom rep inc description of target
#------------------- ---------- -------------------- ---------- --------- ------ ----- --------- ------ ----- --- --- --- --- --- --- --- --- ---------------------
GLGNCOCG_00164 - K00001 - 1.3e-52 177.0 0.4 1.5e-52 176.8 0.4 1.0 1 0 0 1 1 1 1 NADP-dependent alcohol dehydrogenase C
GLGNCOCG_00550 - K00001 - 8.8e-48 161.1 1.0 9.8e-48 160.9 1.0 1.0 1 0 0 1 1 1 1 Alcohol dehydrogenase
GLGNCOCG_02093 - K00001 - 1.1e-05 22.2 3.7 1.3e-05 21.9 3.7 1.1 1 0 0 1 1 1 1 NAD(P) transhydrogenase subunit alpha part 1
GLGNCOCG_00799 - K00001 - 0.00063 16.4 1.5 0.16 8.4 0.3 2.2 2 0 0 2 2 2 2 Glutamate synthase [NADPH] small chain
GLGNCOCG_01761 - K00001 - 0.0011 15.6 0.9 0.0019 14.8 0.3 1.6 2 0 0 2 2 2 1 Alanine dehydrogenase 2
GLGNCOCG_01061 - K00001 - 0.0035 13.9 0.0 0.0057 13.2 0.0 1.4 1 0 0 1 1 1 1 UDP-N-acetyl-D-glucosamine 6-dehydrogenase
GLGNCOCG_01051 - K00001 - 0.0053 13.3 0.3 0.0077 12.8 0.3 1.2 1 0 0 1 1 1 1 Glutamate dehydrogenase
与emapper结果比较
## emapper-2.1.9,evalue:default, 0.001;eggnog_5.02_database/eggnog_proteins.dmnd
## /apps/anaconda3/envs/eggnog-mapper-2.1.9/bin/emapper.py -i GCA_002343985.1_ASM234398v1_genomic.faa --cpu 20 -o emapper_out --override --temp_dir .
(base) [yut@node01 TestData]$ awk -F"\t" 'NR>1{print $1,$12}' emapper_out.emapper.annotations|grep ko|grep -v "#"|wc -l
1232
## kofamscan: evalue 1e-3
(base) [yut@node01 TestData]$ time exec_annotation -p ~/Database/KoFamDB20231025/profiles -k ~/Database/KoFamDB20231025/ko_list --cpu 10 -E 1e-3 -f mapper --no-report-unannotated -o GCA_002343985.1_ASM234398v1_genomic.faa_kofamscan_mapper_annot_format_1e-3.tsv GCA_002343985.1_ASM234398v1_genomic.faa &
(base) [yut@node01 TestData]$ wc -l GCA_002343985.1_ASM234398v1_genomic.faa_kofamscan_mapper_annot_format_1e-3.tsv
1218 GCA_002343985.1_ASM234398v1_genomic.faa_kofamscan_mapper_annot_format_1e-3.tsv
emapper在1e-3的阈值下要比kofamscan在1e-5多16个基因的KO注释,然而当降低kofamscan的evalue到1e-3也仅多了两个基因的KO。
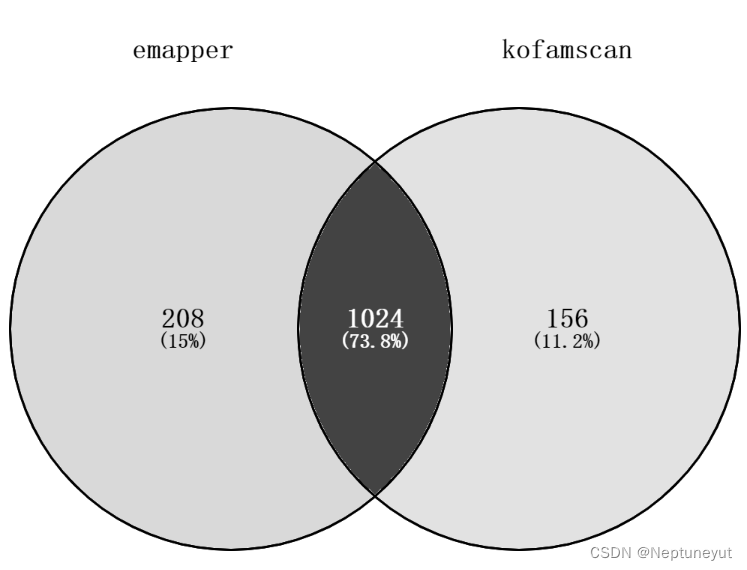
-
对另外多个细菌基因组注释结果统计
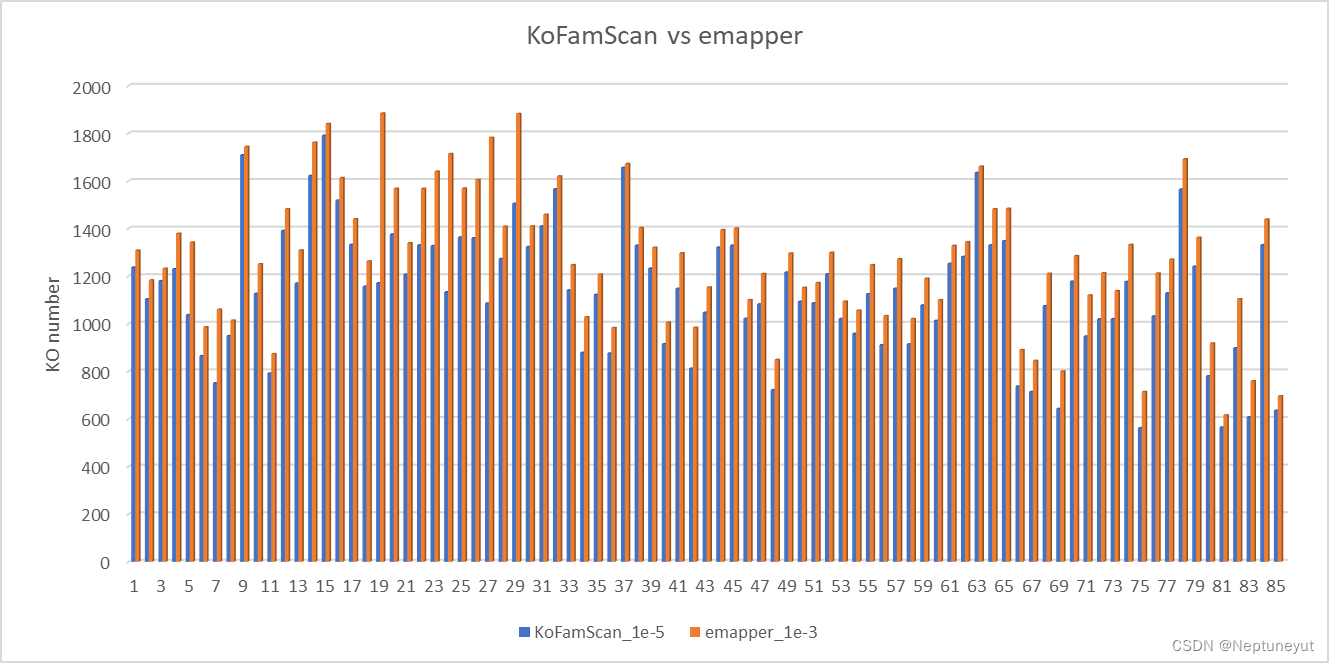
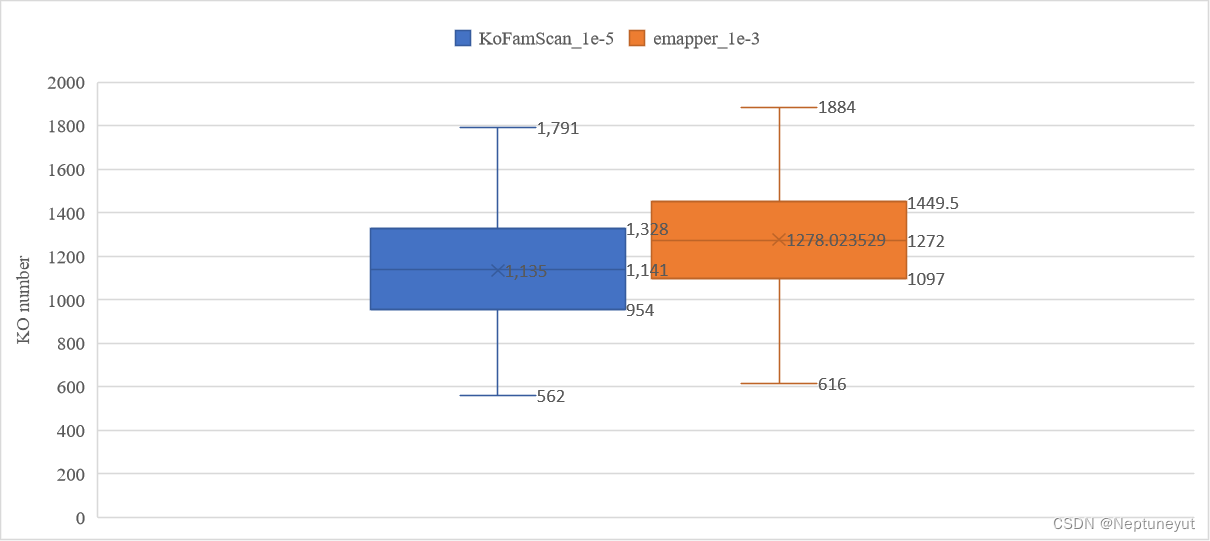
-
85个基因组两种注释的基因数量(包含KO)差异
-
85个基因组两种注释的KO数量差异
其他参数
Usage: exec_annotation [options] <query>
<query> FASTA formatted query sequence file
-o <file> File to output the result [stdout]
-p, --profile <path> Profile HMM database
-k, --ko-list <file> KO information file
--cpu <num> Number of CPU to use [1]
-c, --config <file> Config file
--tmp-dir <dir> Temporary directory [./tmp] # 不写的话默认是当前目录下创建tmp
-E, --e-value <e_value> Largest E-value required of the hits
-T, --threshold-scale <scale>
The score thresholds will be multiplied by this value
-f, --format <format> Format of the output [detail]
detail: Detail for each hits (including hits below threshold) # 按照hit展示,一个基因可能有多个hits,默认格式。显示基因名称、分配的 K 编号、KO 的阈值、hmmsearch 分数和 E 值以及 KO 的定义。此外,如果得分高于阈值,则在行首添加星号 "*"。
detail-tsv: Tab separeted values for detail format # 制表符分隔与上面一样
mapper: KEGG Mapper compatible format # 两列,基因与KO
mapper-one-line: Similar to mapper, but all hit KOs are listed in one line # 一个基因的多个hits在一行
--[no-]report-unannotated Sequence name will be shown even if no KOs are assigned
Default is true when format=mapper or mapper-all,
false when format=detail
--create-alignment Create domain annotation files for each sequence
They will be located in the tmp directory
Incompatible with -r
-r, --reannotate Skip hmmsearch
Incompatible with --create-alignment
--keep-tabular Neither create tabular.txt nor delete K number files
By default, all K number files will be combined into
a tabular.txt and delete them
--keep-output Neither create output.txt nor delete K number files
By default, all K number files will be combined into
a output.txt and delete them
Must be with --create-alignment
-h, --help Show this message and exit
参考
Kofamscan
KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold
IF: 5.8 Q1 B3